NetMHCPan, a widely used tool for predicting peptide binding to major histocompatibility complex (MHC) molecules, is essential for understanding immune responses. However, the tool’s binaries are currently available only for the x86_64 architecture, whether on Darwin (macOS) or Linux. As I intended to conduct test runs on my Apple Silicon device (arm64), I encountered the following error:
1netMHCpan: no binaries found for Darwin_arm64 /net/sund-nas.win.dtu.dk/storage/services/www/packages/netMHCpan/4.1/netMHCpan-4.1/Darwin_arm64/bin/netMHCpan
To address this, one option is to run NetMHCPan using Rosetta, a dynamic binary translator that translates executable code on-the-fly. In simple terms, Rosetta intercepts instructions intended for one architecture (Intel, in this case) and converts them into instructions compatible with another architecture (Apple Silicon).
Ensure that Rosetta is installed on your machine. If not, use the command softwareupdate --install-rosetta to install it.
Let’s walk through the steps of setting up NetMHCPan on Apple Silicon.
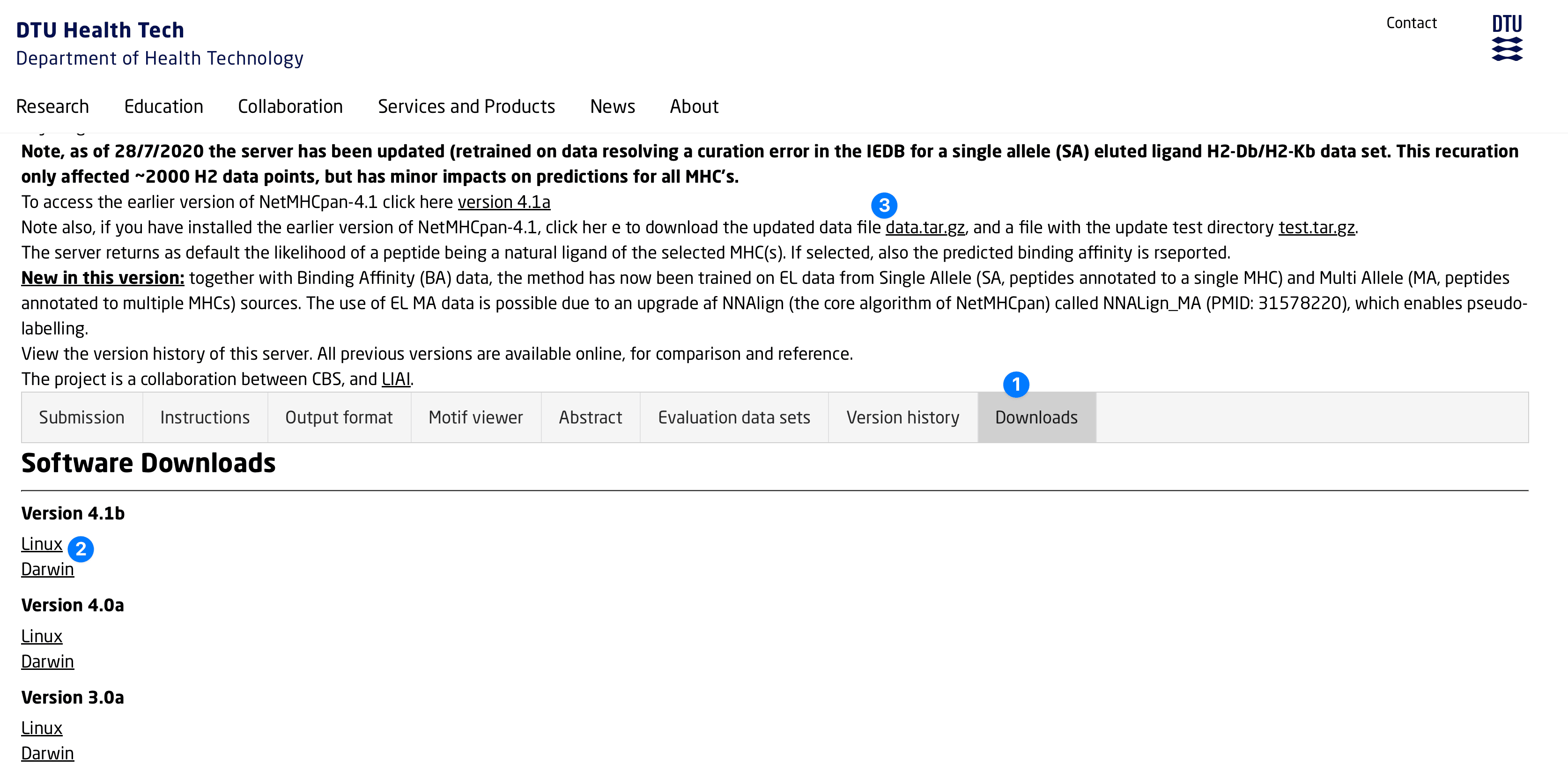
Obtain Binaries and Data
- Visit the NetMHCPan website and navigate to the Downloads tab.
- Download either
netMHCpan-4.1b.Darwin.tar.gzornetMHCpan-4.1b.Linux.tar.gz, depending on your platform - Download
data.tar.gz
Organise Binaries and Data
Extract both the binary and data compressed files into a new folder, for instance, ~/Tools/netMHCpan. Ensure to correctly place the data folder in the designated location.
1mkdir ~/Tools/netMHCpan
2cd ~/Tools/netMHCpan
3
4tar -xvf netMHCpan-4.1b.Darwin.tar.gz
5tar -xvf data.tar.gz
Move the data into the netMHCpan/netMHCpan-4.1 folder.
1mv data netMHCpan-4.1/
2cd ~/Tools/netMHCpan/netMHCpan-4.1/Darwin_x86_64/
Here is the tree structure of the ~/Tools/netMHCpan folder:
1firas@MBP ~/Tools/netMHCpan took 458ms 2024-01-22 20:07:34
2❯ tree -L 3
3.
4└── netMHCpan-4.1
5 ├── Darwin_x86_64
6 │ ├── bin
7 │ └── data -> ../data
8 ├── data
9 │ ├── B0702.fsa
10 │ ├── MHC_pseudo.dat
11 │ ├── README
12 │ ├── all_varcontacts.nlist
13 │ ├── allelenames
14 │ ├── matrices
15 │ ├── synlist
16 │ ├── synlist.bin
17 │ ├── synlist_netMHCpan4.1
18 │ ├── synlist_netMHCpan4.1b
19 │ ├── threshold
20 │ ├── training.pseudo
21 │ └── version
22 ├── netMHCpan
23 ├── netMHCpan-4.1.readme
24 ├── netMHCpan.1
25 └── test
26 ├── B0702.fsa
27 ├── NetMHCpan_out.xls
28 ├── comm
29 ├── test.fsa
30 ├── test.fsa.out
31 ├── test.pep
32 ├── test.pep.out
33 ├── test.pep_BA.out
34 ├── test.pep_userMHC.out
35 └── test.pep_wBA.out
Modify for Apple Silicon
Now, edit the file ~/Tools/netMHCpan/netMHCpan-4.1/netMHCpan to change the following three lines:
1setenv NMHOME '~/Tools/netMHCpan/netMHCpan-4.1'
2setenv UNIX 'Darwin'
3setenv AR 'x86_64'
In essence, we are specifying the current location of the netMHCpan folder on our system and then hard-coding the X86_64 architecture by disabling the checks.
Here is the fully edited version:
1#! /bin/tcsh -f
2
3# This the main NetMHCpan 4.1 script. It only acts as the frontend to the
4# software proper, a compiled binary.
5#
6# VERSION: 2019 Dec 9 launch
7#
8
9###############################################################################
10# GENERAL SETTINGS: CUSTOMIZE TO YOUR SITE
11###############################################################################
12
13# full path to the NetMHCpan 4.0 directory (mandatory)
14#setenv NMHOME /net/sund-nas.win.dtu.dk/storage/services/www/packages/netMHCpan/4.1/netMHCpan-4.1
15setenv NMHOME '~/Tools/netMHCpan/netMHCpan-4.1'
16
17# determine where to store temporary files (must be writable to all users)
18
19if ( ${?TMPDIR} == 0 ) then
20 setenv TMPDIR /tmp
21endif
22
23# determine platform (do not change this unless you don't have 'uname'!)
24setenv UNIX 'Darwin' #`uname -s`
25setenv AR 'x86_64' #`uname -m`
26
27###############################################################################
28# NOTHING SHOULD NEED CHANGING BELOW THIS LINE!
29###############################################################################
30
31# other settings
32set PLATFORM = `echo $UNIX $AR | awk '{print $1"_"$2}'`
33setenv NETMHCpan $NMHOME/$PLATFORM
34setenv DTUIBSWWW www
35setenv NetMHCpanWWWPATH /services/NetMHCpan/tmp/
36setenv NetMHCpanWWWDIR /usr/opt/www/pub/CBS/services/NetMHCpan/tmp
37
38# main ========================================================================
39if ( -x $NETMHCpan/bin/netMHCpan ) then
40 $NETMHCpan/bin/netMHCpan $*
41else
42 echo netMHCpan: no binaries found for $PLATFORM $NETMHCpan/bin/netMHCpan
43endif
44
45# end of script ===============================================================
With these adjustments, NetMHCPan can be seamlessly utilised for analysis on the Apple Silicon architecture.